(C) 2012 Stephen Stern. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Solanum is one of the largest plant genera and exhibits a wide range of morphological diversity. Solanum section Gonatotrichum, the focus of this study, is unique within the genus because of its fruits that swell with turgor pressure and explosively dehisce to disperse the seeds. We infer phylogenetic relationships within section Gonatotrichum using DNA sequence data from two nuclear regions (ITS and the granule-bound starch synthase gene [GBSSI or waxy]) and the chloroplast region trnT-F. The resulting phylogenetic trees support the monophyly of the section with the inclusion of Solanum lignescens, a species not previously thought to belong to the group due to the presence of stellate hairs. This inclusion of this species in section Gonatotrichum suggests that the simple, often geniculate hairs of species in the group may represent reduced stellate hairs. The presence of heterantherous flowers appears to be derived in the section, but this character is largely lost in Solanum parcistrigosum.
explosive fruit dehiscence, Neotropics, Solanum, phylogeny
Solanum (Solanaceae), with approximately 1500 species, is one of the 10 largest flowering plant genera (
Solanum section Gonatotrichum is morphologically unusual within the Brevantherum clade because the species traditionally placed within it have simple rather than stellate hairs and unique fruits with explosive dehiscence (see below;
Species in section Gonatotrichum are native to North, Central, and South America. They are herbs or small, woody shrubs with short inflorescences and simple hairs (except Solanum lignescens, which has stellate pubescence; see below). Flowers in the section have corollas ranging from 1–2.5 cm in diameter. The largest flowers are those of Solanum turneroides and Solanum evolvuloides, which also exhibit marked heteranthery, in which one filament is nearly double the length of the other four. The fruits of species in section Gonatotrichum are unique within the genus. They have a thin pericarp with a watery mesocarp held under pressure until they explosively dehisce. The fruits are white, yellow, or green, nearly transparent, and turgid before explosive dehiscence and deflated and shriveled after dehiscence.
Some species within section Gonatotrichum are relatively widespread (Solanum deflexum, Solanum parcistrigosum, and Solanum turneroides), whereas others are narrowly distributed and relatively inconspicuous, making them among the least collected species of Solanum. As
Species placed in section Gonatotrichum have been subjected to two previous phylogenetic studies.
In this paper we use molecular phylogenetic methods to 1) examine the phylogenetic relationships of section Gonatotrichum with other members of the genus, 2) test the monophyly of section Gonatotrichum, 3) test the monophyly of species within the section and examine selected species-level relationships, and 4) examine geographical distributions and morphological patterns within the section.
Materials and methods Taxon samplingSeven of the eight recognized species of section Gonatotrichum were sampled. We were unable to obtain high quality genomic DNA for Solanum hoffmanseggii, an undercollected species from Amazonian Brazil. Multiple accessions were sampled for four species, with four accessions sampled for Solanum parcistrigosum, and two each for Solanum evolvuloides, Solanum manabiense, and Solanum deflexum. In addition to the species of section Gonatotrichum, we included members of Solanum as outgroups guided by results from previous studies showing other members of the Brevantherum clade to be sister to the section (
Total genomic DNA was extracted from fresh, silica gel-dried, or herbarium material using the DNeasy plant mini extraction kit (Qiagen, Inc., Valencia, California). PCR amplification for each gene region followed standard procedures described in
PCR products were cleaned using the Promega Wizard SV PCR Clean-Up System (Promega Corporation, Madison, Wisconsin). The University of Utah DNA Sequencing Core Facility performed sequencing on an ABI automated sequencer. Sequences were edited in Sequencher (Gene Codes Corp., Ann Arbor, Michigan) and all new sequences were submitted to GenBank; accession numbers are listed in Appendix 1.
Sequence alignment and analysisSequence alignment for all of the gene regions was straightforward and performed visually using Se-Al (
Parsimony analyses were performed on each dataset separately and on the combined dataset using PAUP*4.0b10 (
Prior to Bayesian analyses, a general model of nucleotide evolution was selected for the separate and the combined datasets using the AIC criterion identified in Modeltest 3.7 (
The parsimony strict consensus and Bayesian majority rule consensus trees of all datasets differed only in the degree of resolution, with Bayesian tree topologies more resolved than parsimony trees (Table 1). Clades with low posterior probabilities, typically those below 0.90 PP but occasionally those with up to 1.0 PP in Bayesian analyses were often collapsed in parsimony strict consensus trees. Descriptive statistics for individual and combined genes are provided (Table 1). More nodes were strongly supported by combining the data than were obtained in any of the separate analyses.
Descriptive statistics for each data set analyzed. Strongly supported nodes for parsimony indicate those with ≥ 90% BS; Bayesian strongly supported nodes are those with ≥ 0.95 PP.
| Data Partition | Aligned Sequence Length | # Parsimony Informative Characters | # MP Trees | Tree Length | CI | RI | # Strongly Supported Nodes Parsimony | Model Selected | # Strongly Supported Nodes Bayesian |
| ITS | 709 | 127 | 14 | 435 | 0.632 | 0.683 | 4 | GTR+I+G | 15 |
| waxy | 2090 | 173 | 40 | 403 | 0.893 | 0.942 | 10 | TIM+G | 21 |
| trnT-F | 1953 | 71 | >135, 000 | 178 | 0.944 | 0.960 | 8 | TVM+G | 12 |
| combined | 4752 | 371 | 14 | 1031 | 0.779 | 0.847 | 13 | GTR+I+G | 25 |
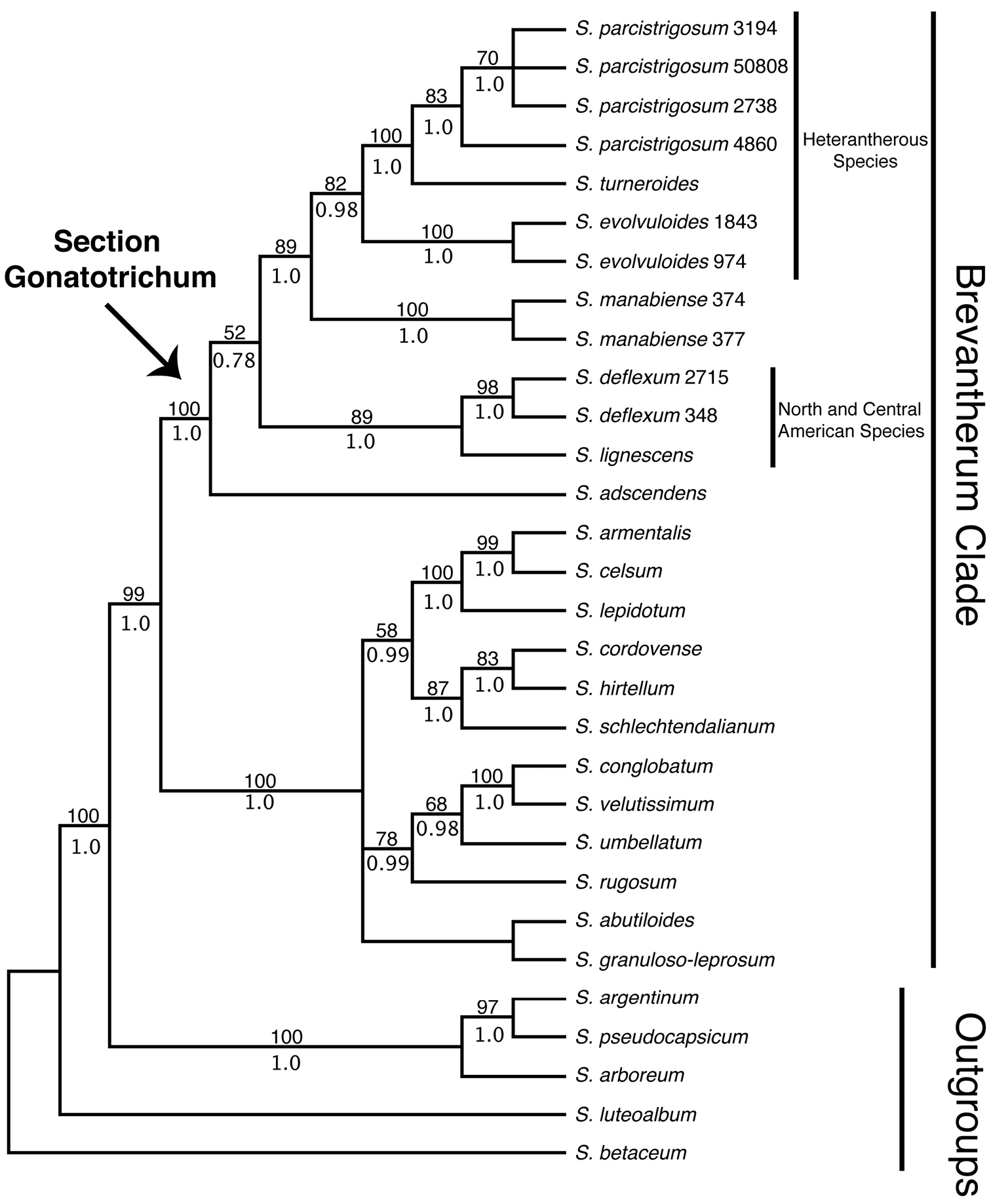
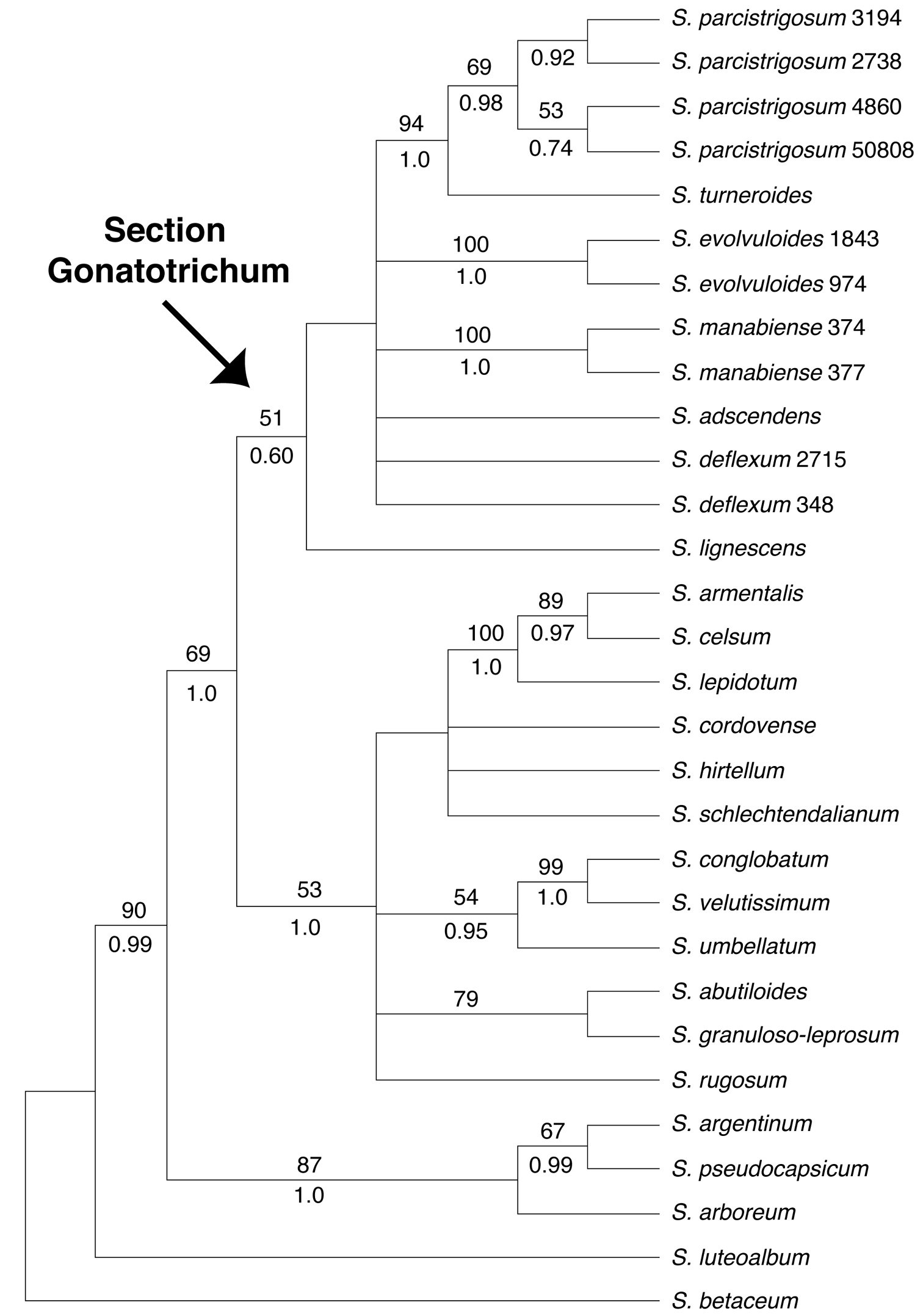
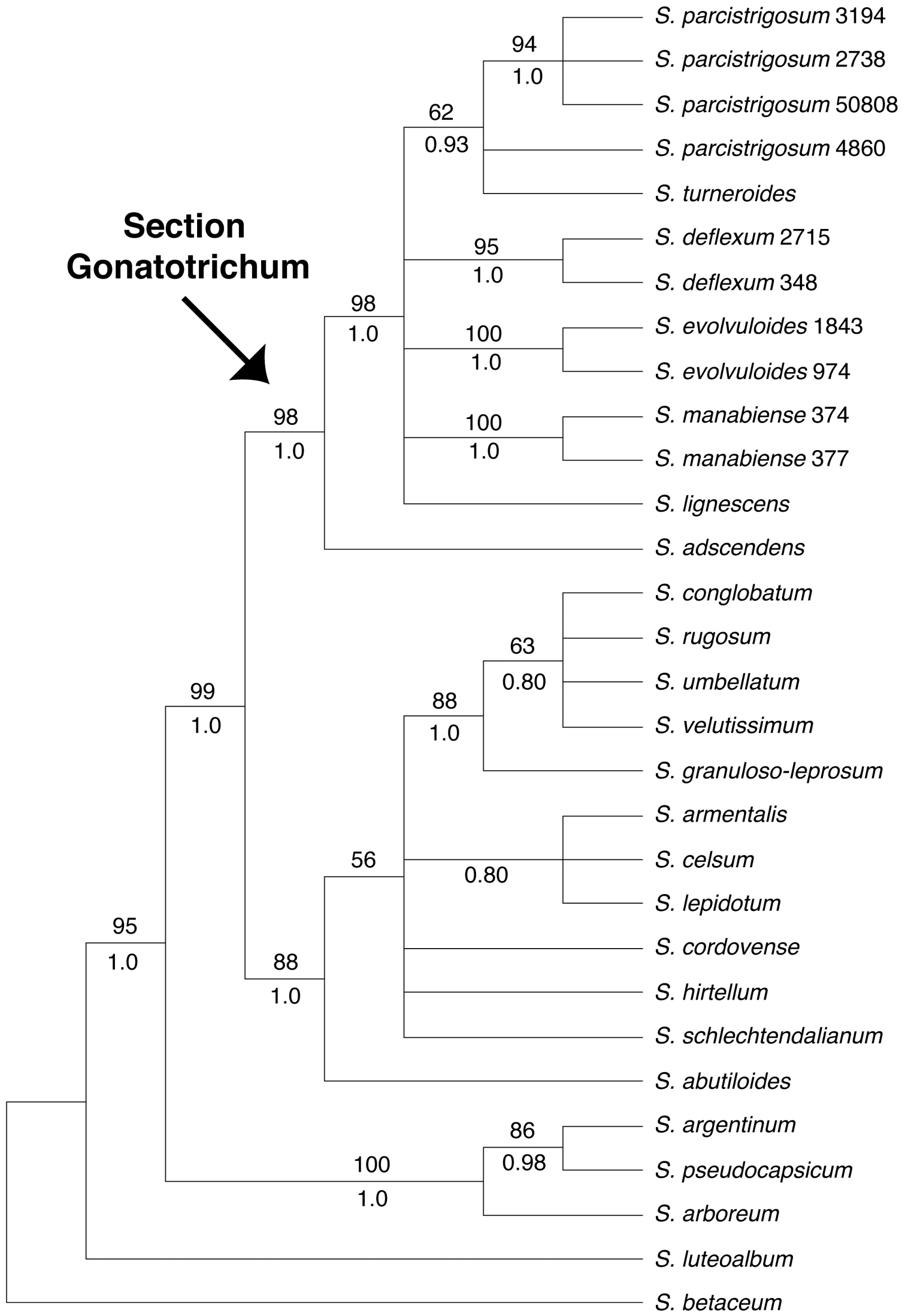
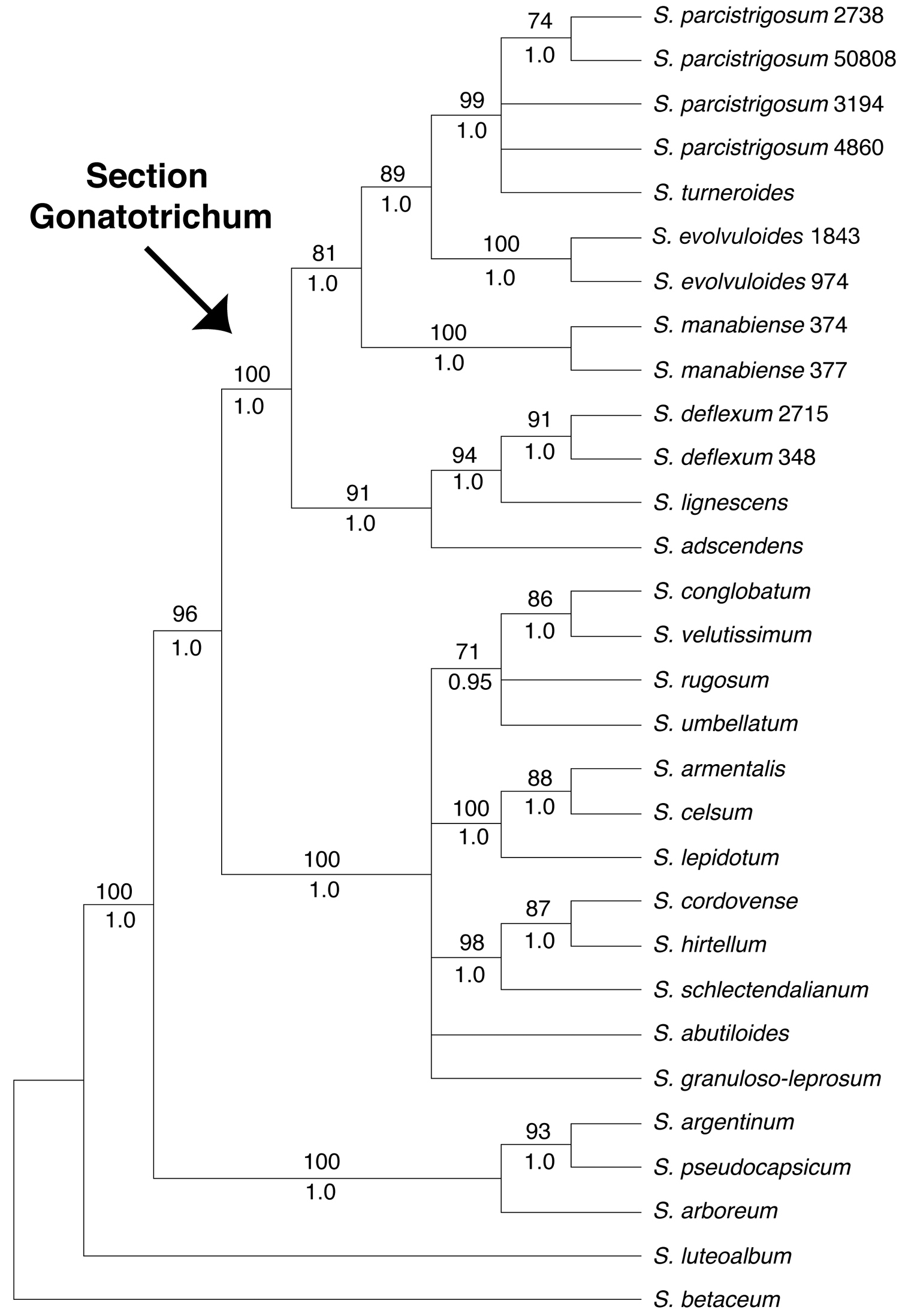
Our discussion will largely be based on the parsimony strict consensus tree of the combined data set, which is a conservative hypothesis of phylogenetic relationships, but areas of the tree that receive strong support in the Bayesian analysis that are less strongly supported in the parsimony analysis will be noted (Fig. 1). The parsimony strict consensus trees for the individual markers are also presented (Fig. 2–4). In parsimony analyses, each DNA sequence region consistently identified the same major, well-supported groups corresponding to the Brevantherum clade and section Gonatotrichum comprising identical species, but relationships within these major clades were often not strongly supported (BS values < 90 %), or were unresolved, and thus cannot be considered conflicting under
Strict consensus of the 14 most parsimonious trees from the concatenated MP analysis of ITS, trnT-F, and waxy data. Bootstrap values > 50% and posterior probabilities are shown above and below the branches, respectively. Numbers after species names indicate collector numbers listed in Appendix 1 for multiple accessions of a single species.
Strict consensus of the 14 most parsimonious trees from the MP analysis of the ITS dataset. Bootstrap values > 50% and posterior probabilities are shown above and below the branches, respectively.
Strict consensus of the more than 135, 000 most parsimonious trees from the MP analysis of the trnT-F dataset. Bootstrap values > 50% and posterior probabilities are shown above and below the branches, respectively.
Strict consensus of the 40 most parsimonious trees from the MP analysis of the waxy (GBSSI) dataset. Bootstrap values > 50% and posterior probabilities are shown above and below the branches, respectively.
Solanum section Gonatotrichum emerges as monophyletic in all analyses and strongly supported in all except the ITS-only tree. The section is strongly supported (99% BS, 1.0 PP in the combined tree) as sister to the remainder of the species sampled from the Brevantherum clade, which form a monophyletic group in all analyses (100% BS, 1.0 PP in the combined tree; for sampling of Brevantherum clade see Fig. 1). Species with duplicate accessions sequenced (Solanum parcistrigosum, Solanum evolvuloides, Solanum manabiense, and Solanum deflexum) were all monophyletic in the combined tree. Within section Gonatotrichum, Solanum turneroides is strongly supported as sister to Solanum parcistrigosum in the combined tree (100% BS, 1.0 PP), and Solanum evolvuloides is sister to this clade (82% BS, 0.98 PP). Solanum manabiense from coastal Ecuador is resolved as sister to this clade (89% BS, 1.0 PP). Solanum deflexum and Solanum lignescens form a clade (89% BS, 1.0 PP). The final species, Solanum adscendens, is sister to the remainder of the species of section Gonatotrichum in the combined tree, although this relationship is poorly supported (52% BS, 0.78 PP) and, as noted above, is not recovered in the ITS and waxy trees.
DiscussionOur data, like those of previous studies, show that Solanum section Gonatotrichum belongs to the Brevantherum clade (sensu
These results indicate that section Gonatotrichum forms a monophyletic group including the Mesoamerican Solanum lignescens. This species had not been considered to be a member of the section Gonatotrichum in previous taxonomic treatments (
Hairs have been used extensively for identification of species and sections in Solanum and have their own standardized terminology in the genus (see
We sampled multiple accessions per taxon in four of the seven species of section Gonatotrichum included in the phylogeny (Solanum evolvuloides, Solanum deflexum, Solanum manabiense, and Solanum parcistrigosum). In the combined and waxy trees, all four species were monophyletic. The four accessions of Solanum parcistrigosum were not monophyletic in the trnT-F tree, and the two accessions of Solanum deflexum did not form a clade in the ITS tree. However, neither of these cases of species non-monophyly were strongly supported. It appears that species limits, at least within these four taxa, are fairly distinct.
Within section Gonatotrichum, the phylogeny from the combined data exhibits clear geographic patterns. The species ranging from North to Central America (Solanum lignescens and Solanum deflexum) form a strongly supported clade, as do the southern South American species (Solanum evolvuloides, Solanum parcistrigosum, and Solanum turneroides) with the Ecuadorian Solanum manabiense sister to the southern South American species. The position of the Brazilian Solanum adscendens as sister to the rest of the clade in the combined and trnT-F trees does not fit this biogeographic pattern and is confounded by the incongruence between the trnT-F and waxy datasets indicated above, although neither of these datasets place Solanum adscendens with the remainder of the southern South American species. Future molecular studies should attempt to include Solanum hoffmanseggii, a poorly known species from Amazonian Brazil and the only member of section Gonatotrichum for which sequence data are not available. Based on morphology, particularly the geniculate hairs that lay flat along the stem, the similar sized flowers and fruits, and the overall distribution, we speculate that Solanum hoffmanseggii will likely be closely related to the southern South American species Solanum parcistrigosum. Sequence data is needed to confirm this relationship and determine its phylogenetic affinities to the rest of the South American taxa of the clade.
Although buzz pollination is virtually universal in Solanum, heteranthery is an unusual trait in the genus and has been shown to have evolved multiple times independently within it (
Future work on section Gonatotrichum should include sequencing of more gene regions and species accessions, especially targeting Solanum hoffmanseggii and Solanum adscendens, to clarify biogeographic patterns in the group. Nothing is known about chromosome numbers or potential hybridization among taxa, and the function of heteranthery in pollination of several species within the group is unclear. Detailed studies of the development and morphology of hairs in Solanum and in section Gonatotrichum in particular may reveal that simple hairs may have arisen by two different evolutionary pathways, either from plesiomorphically simple hairs or by reduction from branched hairs. Finally, more in-depth studies of the entire Brevantherum clade are needed to clarify its species limits, phylogenetic relationships, and morphological and biogeographical patterns.
We thank E. Tepe, K. Leo, and T. Weese for laboratory assistance, J. Stehmann and L. Giacomin for help with Brazilian specimens, E. Tepe and M. Nee for field assistance, S. Knapp for advice on Solanum lignescens, and S. Knapp and two anonymous reviewers for improving the manuscript. This work was supported by NSF grant DEB-0316614 to L.B.
Summary of species, collection location, vouchers, herbarium acronym, and GenBank accession numbers for taxa used in this study provided in the order ITS, trnT-F, and waxy. BIRM samples have the seed accession number for the Solanaceae collection at the University of Birmingham, UK (now transferred to the Botanic Garden of the University of Nijmegen, Nijmegen, The Netherlands, http://www.bgard.science.ru.nl/solanaceae/).
| Species | Collection Location | Voucher and Herbarium Acronym | GenebankAccession Number for ITS | Genebank Accession Number for trnT-F | GenBank Accession Number for waxy |
|---|---|---|---|---|---|
| Solanum abutiloides (Griseb.) Bitter & Lillo | BIRM S.0655 | Olmstead S-73 (WTU); | AF244716 | AY266236 | AY562948 |
| Solanum adscendens Sendtn. | Brazil | Stehmann 6001 (BHCB) | JN542580 | JN661818 | JN661835 |
| Solanum arboreum Dunal | Costa Rica | Bohs 2521 (UT) | AF244719 | DQ180424 | AY996381 |
| Solanum argentinum Bitter & Lillo | Argentina | Bohs 2539 (UT) | AF244718 | DQ180425 | AY996382 |
| Solanum armentalis J.L. Gentry & D’Arcy | Costa Rica | Bohs 2593 (UT); | JN542581 | JN661819 | JN661836 |
| Solanum betaceum Cav. | Bolivia | Bohs 2468 (UT) | AF244713 | DQ180426 | AY996387 |
| Solanum celsum Standl. & C.V. Morton | Costa Rica | Bohs 2592 (UT) | JN542582 | JN661820 | JN661837 |
| Solanum conglobatum Dunal | Bolivia | Bohs 2740 (UT) | JN542583 | JN661821 | JN661838 |
| Solanum cordovense Sessé & Moc. | Costa Rica | Bohs 2693 (UT) | AF244717 | DQ180480 | AY996401 |
| Solanum deflexum Greenm. | Costa Rica | Bohs 2715 (UT) | JN542584 | DQ180427 | DQ169025 |
| Solanum deflexum Greenm. | Mexico | Smith & Rojas 348 (NY) | JN542585 | JN661822 | JN661839 |
| Solanum evolvuloides Giacomin & Stehmann | Brazil | Jardim 1843 (NY) | JN542586 | JN661823 | JN661840 |
| Solanum evolvuloides Giacomin & Stehmann | Brazil | Giacomin 974 (BHCB) | JN542587 | JN661824 | JN661841 |
| Solanum granuloso-leprosum Dunal | Paraguay | Bohs 3190 (UT) | JN542588 | JN661825 | JN661842 |
| Solanum hirtellum (Spreng.) Hassl. | Paraguay | Bohs 3166 (UT) | JN542589 | JN661826 | JN661843 |
| Solanum lepidotum Dunal | Costa Rica | Bohs 2621 (UT) | JN542590 | DQ180486 | DQ169035 |
| Solanum lignescens Fernald | Mexico | Linares 3536 (MEXU) | JN542591 | JN661827 | JN661844 |
| Solanum luteoalbum Pers. | BIRM S.0042 | Bohs 2337 (UT) | AF244715 | DQ180433 | AY562957 |
| Solanum manabiense S. Stern | Ecuador | Stern & Tepe 377 (UT) | JN542592 | JN661828 | JN661845 |
| Solanum manabiense S. Stern | Ecuador | Stern & Tepe 374 (UT) | JN542593 | JN661829 | JN661846 |
| Solanum parcistrigosum Bitter | Bolivia | Bohs 2738 (UT) | JN542595 | DQ180431 | DQ169013 |
| Solanum parcistrigosum Bitter | Argentina | Nee & Bohs 50808 (NY) | JN542597 | JN661832 | JN661849 |
| Solanum parcistrigosum Bitter | Paraguay | Bohs 3194 (UT) | JN542594 | JN661830 | JN661847 |
| Solanum parcistrigosum Bitter | Brazil | Aparecida et al. 4860 (NY) | JN542596 | JN661831 | JN661848 |
| Solanum pseudocapsicum L. | BIRM S.0870 | no voucher | AF244720 | DQ180436 | AY562963 |
| Solanum rugosum Dunal | Costa Rica | Bohs 3011 (UT) | JN542598 | DQ180490 | DQ169046 |
| Solanum schlechtendalianum Walp. | Costa Rica | Bohs 2915 (UT) | JN542599 | DQ180491 | DQ169047 |
| Solanum turneroides Chodat | Bolivia | Nee 51716 (NY); | JN542600 | DQ180439 | DQ169051 |
| Solanum umbellatum Mill. | Costa Rica | Bohs 2560 (UT) | JN542601 | JN661833 | JN661850 |
| Solanum velutissimum Rusby | Bolivia | Nee 51780 (NY) | JN542602 | JN661834 | JN661851 |
Aligned dataset. (doi: 10.3897/phytokeys.8.2199.app) File format: NEXUS matrix file.
Explanation note: Aligned dataset; alignment performed visually using Se-Al (Rambaut 1996).
Copyright notice: This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.