|
||
|
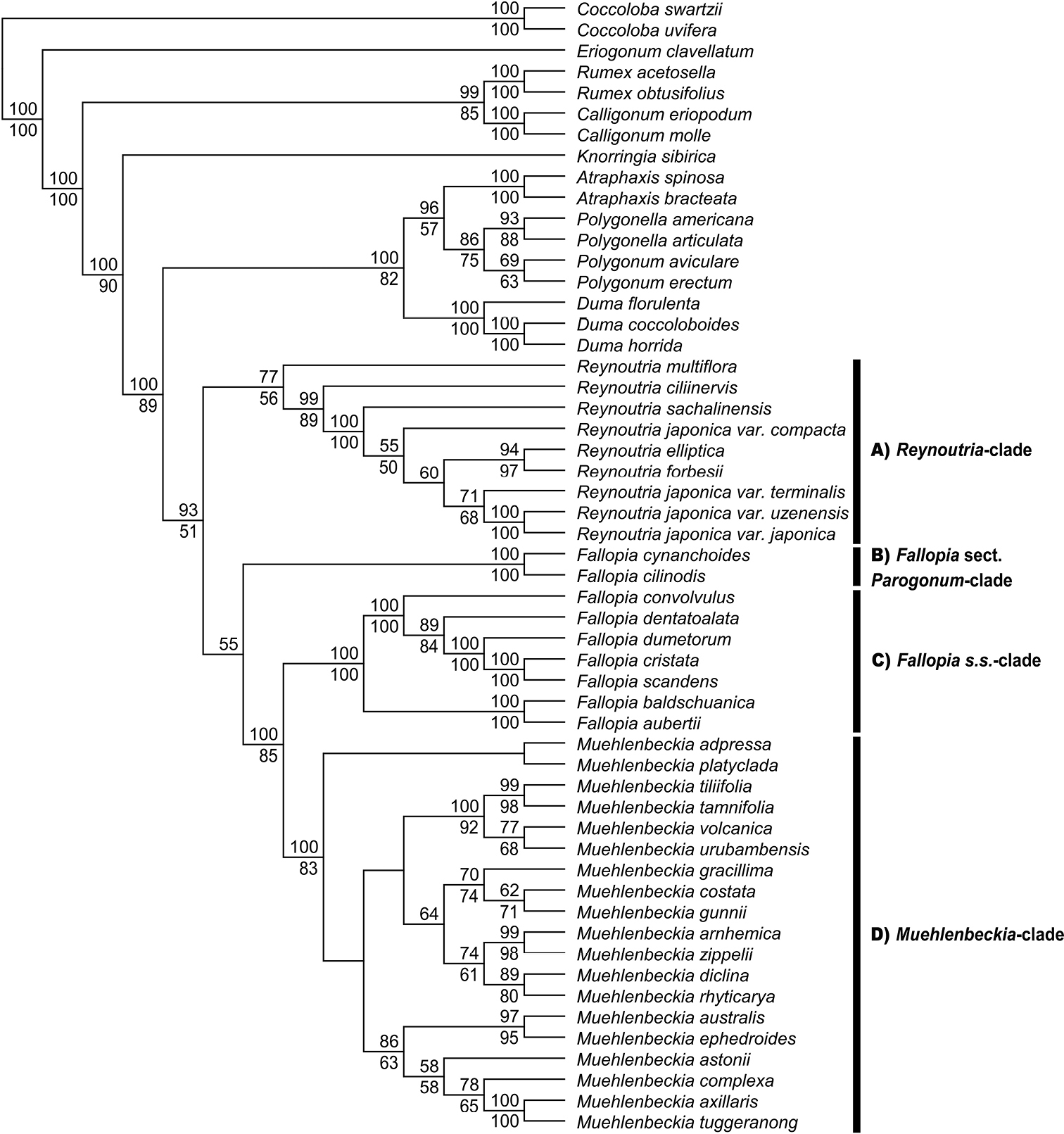
A total evidence phylogenetic tree generated by a Maximum Likelihood analysis of concatenated nuclear (ITS and LEAFYi2) and chloroplast (matK, rbcL, rps16-trnK and trnL-trnF) sequence data. Bootstrap support values (≥ 50%) are displayed above and below the nodes for Maximum Likelihood and Maximum Parsimony analyses, respectively. Maximum Parsimony analysis recovered eight equally parsimonious trees (3099 steps). The main clades within subtribe Reynoutriinae are marked with bars. |