|
||
|
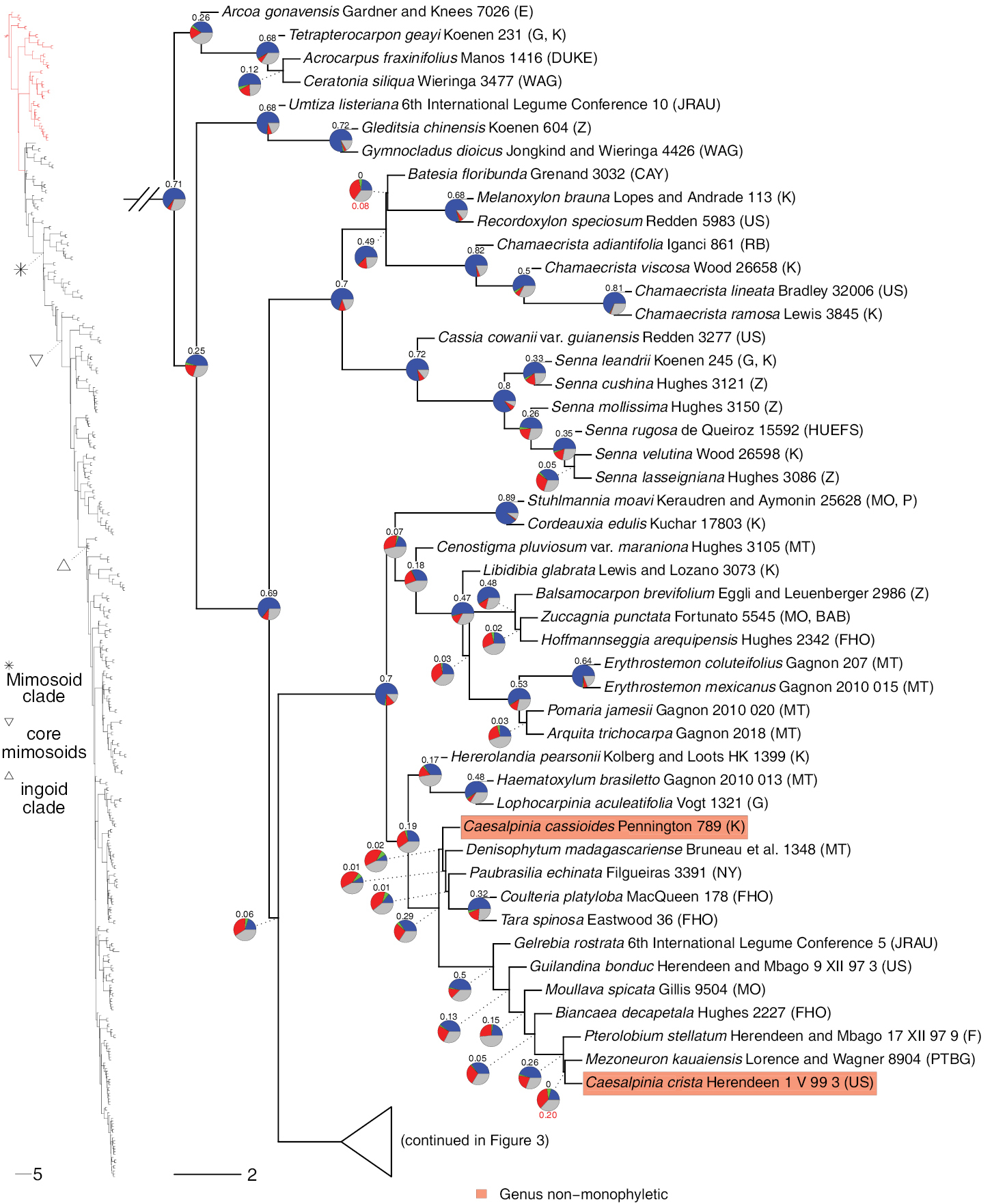
Phylogeny of Caesalpinioideae, part 1 (continued in Figs 3–12). Left part of figure shows complete Caesalpinioideae phylogeny with highlighted in red the part shown in detail on the right. Depicted phylogeny is the ASTRAL (Zhang et al. 2018) phylogeny based on 821 single-copy nuclear gene trees, with branch lengths expressed in coalescent units and terminal branches assigned an arbitrary uniform length for visual clarity. Genera resolved as (potentially) non-monophyletic are highlighted and clades recognised by Koenen et al. (2020b) are labelled. Support for relationships is based on gene tree conflict: pie charts show the fractions of supporting and conflicting gene trees per node calculated using PhyParts (Smith et al. 2015), with blue representing supporting gene trees, green gene trees supporting the most common alternative topology, red gene trees supporting further alternative topologies and grey gene trees uninformative for this node. Numbers above nodes are Extended Quadripartition Internode Certainty scores calculated with QuartetScores (Zhou et al. 2020). Numbers below nodes are the outcome of ASTRAL’s polytomy test (Sayyari and Mirarab 2018), which tests for each node whether the polytomy null model can be rejected. Only non-significant (i.e. > 0.05) scores are shown, i.e. only for nodes that are better regarded as polytomies according to the test. |