|
||
|
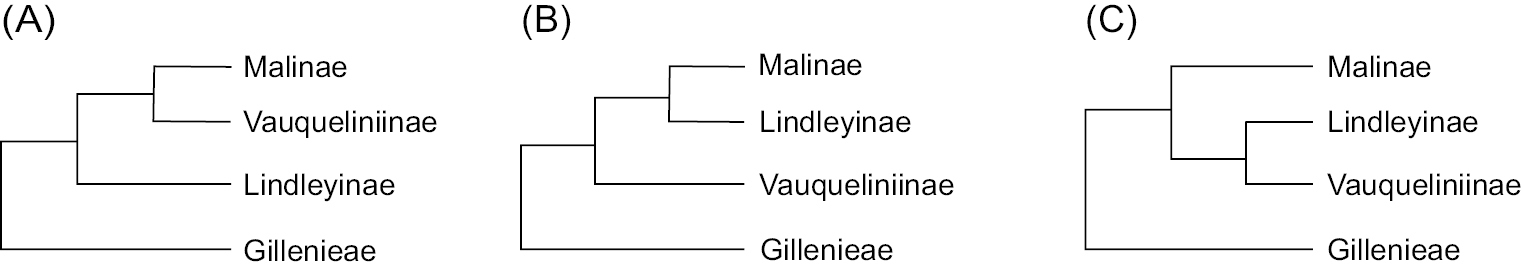
Phylogenetic hypotheses among subtribes within the apple tribe Maleae. A plastome-based topology (current study; Liu et al. 2020a); 11 plastid regions- and nuclear ribosomal internal transcribed spacer (nrITS)-based topology (atpB-rbcL, psbA-trnH, rbcL, rpl16 intron, rpl20-rps12, rps16 intron, trnC-ycf6, trnG-trnS, trnH-rpl2, trnL-trnF, and trnK + matK; Lo and Donoghue 2012); transcriptome-based topology (Xiang et al. 2017; Zhang et al. 2023) B single-copy nuclear genes (SCN genes)-based topology (inferred from Maximum Likelihood (ML) methods: Liu et al. 2022; Jin et al. 2023) C SCN genes-based topology (ASTRAL-III species tree: Liu et al. 2022; Jin et al. 2023). |