Citation: Cochran AT, Prado J, Schuettpelz E (2014) Tryonia, a new taenitidoid fern genus segregated from Jamesonia and Eriosorus (Pteridaceae). PhytoKeys 35: 23–43. doi: 10.3897/phytokeys.35.6886
The Neotropical fern genera Eriosorus and Jamesonia have long been thought of as close relatives. Molecular phylogenetic studies have confirmed this notion but have also revealed that neither genus is monophyletic with respect to the other. As a result, all known species of Eriosorus were recently subsumed under the older generic name Jamesonia. Here, through an analysis of a four-gene plastid dataset, we show that several species traditionally treated in Eriosorus are in fact more closely related to other taenitidoid fern genera (namely Austrogramme, Pterozonium, Syngramma, and Taenitis) than they are to the large Jamesonia sensu lato clade. Tryonia Schuettp., J.Prado & A.T.Cochran gen. nov. is described to accommodate these species and four new combinations are provided. Tryonia is confined to southeastern Brazil and adjacent Uruguay; it is distinct (from most species of Jamesonia) in having stramineous rachises.
Brazil, phylogeny, pteridophytes, Taenitidoideae, taxonomy
The Neotropical genus Jamesonia Hook. & Grev. sensu stricto is among the most distinctive of all fern genera. It has linear, indeterminate leaves bearing highly reduced, coriaceous pinnae covered with dense pubescence (
Jamesonia pulchra Hook. & Grev., the type species of Jamesonia. Ewan 16100 (US), inset detail of (castaneous) rachis magnified 4×.
Jamesonia aureonitens (Hook.) Christenh., the type species of Eriosorus. Hutchison 5504 (US), inset detail of (castaneous) rachis magnified 4×.
Jamesonia congesta (Christ) Christenh., a species with generalized morphology (
Although it is clear that species of Jamesonia sensu stricto are intermixed with those previously assigned to Eriosorus, relationships remain rather poorly supported and additional studies are needed to better resolve the evolutionary history of this group. With that said, the isolated phylogenetic position revealed for one Brazilian species requires special attention. In the most comprehensive study of Jamesonia sensu lato to date (
Here, through analyses of a four-gene (atpA, chlL, rbcL, and rps4) plastid dataset that incorporates many Eriosorus and Jamesonia sensu stricto species, as well as a broad sampling of related genera, we aim to better resolve the phylogenetic position of Eriosorus myriophyllus and allied species. Based on our results, we describe a new genus, Tryonia Schuettp., J.Prado & A.T.Cochran, to accommodate this species and its closest allies.
Tryonia myriophylla (Sw.) Schuettp., J.Prado & A.T.Cochran, the type species of Tryonia. Smith 1795 (US), inset detail of (stramineous) rachis magnified 4×.
A total of thirty-eight collections were sampled for the phylogenetic analysis, including four individuals of Eriosorus myriophyllus, nine other species of Eriosorus, eight Jamesonia sensu stricto species, and seventeen additional species representing other genera in the taenitidoid clade (
Collections included in our phylogenetic analyses supporting the recognition of Tryonia, with voucher information and corresponding GenBank accession numbers.
| Species | Voucher | atpA | chlL | rbcL | rps4 | FLDB |
|---|---|---|---|---|---|---|
| Actiniopteris dimorpha Pic.Serm. | Schneider s.n. (GOET) | EF452066 | KJ416295 | EF452130 | KJ416352 | 3515 |
| Actiniopteris semiflabellata Pic.Serm. | Smith s.n. (UC) | KJ416270 | KJ416296 | KJ416326 | KJ416353 | 3742 |
| Anogramma leptophylla (L.) Link | Schuettpelz 1079 (DUKE) | KJ416271 | KJ416297 | KJ416327 | KJ416354 | 4822 |
| Austrogramme decipiens (Mett.) Hennipman | van der Werff 16114 (UC) | NA | NA | NA | AF321702 | NA |
| Austrogramme marginata (Mett.) E.Fourn. | Hodel 1454 (UC) | NA | NA | NA | AY357704 | NA |
| Cosentinia vellea (Aiton) Tod. | Larsson 55 (UPS) | KJ416272 | KJ416298 | KJ416328 | KJ416355 | 8670 |
| Jamesonia alstonii A.F.Tryon | Moran 8248 (DUKE) | KJ416273 | KJ416299 | KJ416329 | KJ416356 | 5587 |
| Jamesonia blepharum A.F.Tryon | Schuettpelz 269 (DUKE) | KJ416274 | KJ416300 | EF452154 | KJ416357 | 2437 |
| Jamesonia brasiliensis Christ | Schuettpelz 1444 (SP) | KJ416275 | KJ416301 | KJ416330 | KJ416358 | 8379 |
| Jamesonia cheilanthoides (Sw.) Christenh. | Rothfels 3964 (DUKE) | KJ416276 | KJ416302 | KJ416331 | KJ416359 | 7694 |
| Jamesonia congesta (Christ) Christenh. | Grusz 08-036 (DUKE) | KJ416277 | KJ416303 | KJ416332 | KJ416360 | 5272 |
| Jamesonia elongata (Grev. & Hook.) J.Sm. | Rothfels 3602 (DUKE) | KJ416278 | KJ416304 | KJ416333 | KJ416361 | 7362 |
| Jamesonia flexuosa (Kunth) Christenh. | Rothfels 08-042 (DUKE) | KJ416279 | KJ416305 | KJ416334 | KJ416362 | 5273 |
| Jamesonia goudotii (Hieron.) C.Chr. | Rothfels 3694 (DUKE) | KJ416280 | KJ416306 | KJ416335 | KJ416363 | 7414 |
| Jamesonia hirta (Kunth) Christenh. | Rothfels 3669 (DUKE) | KJ416281 | KJ416307 | KJ416336 | KJ416364 | 7397 |
| Jamesonia insignis (Kuhn) Christenh. | Salino 3010 (UC) | NA | NA | NA | AF321708 | NA |
| Jamesonia pulchra Hook. & Grev. | Sánchez-Baracaldo 306 (UC) | NA | NA | NA | AF321746 | NA |
| Jamesonia rotundifolia Fée | Sundue 1357 (DUKE) | KJ416282 | KJ416308 | KJ416337 | KJ416365 | 6049 |
| Jamesonia scammaniae A.F.Tryon | Rothfels 2631 (DUKE) | KJ416283 | KJ416309 | KJ416338 | KJ416366 | 5588 |
| Jamesonia verticalis Kunze | Rothfels 3638 (DUKE) | KJ416284 | KJ416310 | KJ416339 | KJ416367 | 7386 |
| Jamesonia warscewiczii (Mett.) Christenh. | Grusz 08-039 (DUKE) | KJ416285 | KJ416311 | KJ416340 | KJ416368 | 5275 |
| Onychium japonicum (Thunb.) Kunze | Schneider s.n. (GOET) | EF452107 | KJ416312 | KJ416341 | NA | 3463 |
| Onychium lucidum (D.Don) Spreng. | Schuettpelz 1161 (DUKE) | KJ416286 | KJ416313 | KJ416342 | NA | 4904 |
| Pityrogramma austroamericana Domin | Schuettpelz 301 (DUKE) | EF452112 | KJ416314 | EF452166 | KJ416369 | 2561 |
| Pityrogramma chaerophylla (Desv.) Domin | Prado 2178 (SP) | KJ416287 | KJ416315 | KJ416343 | KJ416370 | 8755 |
| Pityrogramma jamesonii (Baker) Domin | Moran 7592 (NY) | EF463857 | KJ416316 | EF452167 | KJ416371 | 3769 |
| Pterozonium brevifrons (A.C.Sm.) Lellinger | Schuettpelz 285 (DUKE) | EF452124 | KJ416317 | EF452175 | KJ416372 | 2453 |
| Pterozonium cyclosorum A.C.Sm. | Brewer 1006 (UC) | NA | NA | NA | AF321703 | NA |
| Pterozonium reniforme (Mart.) Fée | Brewer 1005 (UC) | NA | NA | NA | AF321704 | NA |
| Syngramma quinata (Hook.) Carr. | Kessler 2273 (L) | NA | NA | NA | AF321701 | NA |
| Taenitis blechnoides (Willd.) Sw. | Schuettpelz 689 (DUKE) | KJ416288 | KJ416318 | KJ416344 | KJ416373 | 4102 |
| Taenitis interrupta Hook. & Grev. | Schuettpelz 851 (DUKE) | KJ416289 | KJ416319 | KJ416345 | KJ416374 | 4270 |
| Tryonia areniticola (Schwartsb. & Labiak) Schuettp., J.Prado & A.T.Cochran | Prado 2169 (SP) | NA | KJ416320 | KJ416346 | KJ416375 | 8433 |
| Tryonia myriophylla (Sw.) Schuettp., J.Prado & A.T.Cochran | Schuettpelz 1411 (SP) | KJ416290 | KJ416321 | KJ416347 | KJ416376 | 8345 |
| Tryonia myriophylla (Sw.) Schuettp., J.Prado & A.T.Cochran | Schuettpelz 1449 (SP) | KJ416291 | KJ416322 | KJ416348 | KJ416377 | 8384 |
| Tryonia myriophylla (Sw.) Schuettp., J.Prado & A.T.Cochran | Schuettpelz 1461 (SP) | KJ416292 | KJ416323 | KJ416349 | KJ416378 | 8396 |
| Tryonia myriophylla (Sw.) Schuettp., J.Prado & A.T.Cochran | Prado 2186 (SP) | KJ416293 | KJ416324 | KJ416350 | NA | 8753 |
| Tryonia schwackeana (Christ) Schuettp., J.Prado & A.T.Cochran | Schuettpelz 1433 (SP) | KJ416294 | KJ416325 | KJ416351 | KJ416379 | 8367 |
† Fern Lab Database voucher number (see http://fernlab.biology.duke.edu for additional information concerning these collections)
Genomic DNA was typically extracted using a modified CTAB protocol (
Primers utilized in this study supporting the recognition of Tryonia.
| Region | Name | Type | Sequence | Reference |
|---|---|---|---|---|
| atpA | atpA-F1 | Forward | GAATCTGATAATGTTGGGGCTG | This study |
| atpA | atpA-R1 | Reverse | AAACATCTCCNGGATAYGCTTC | This study |
| chlL | chlL-F1 | Forward | GRATTGGMAARTCAACAACTAGCTG | This study |
| chlL | chlL-R1 | Reverse | CBAGTACRGGCATGGGRCAAGCTTC | This study |
| rbcL | ES-rbcL-1F | Forward | ATGTCACCACAAACGGAGACTAAAGC | |
| rbcL | ES-rbcL-1361R | Reverse | TCAGGACTCCACTTACTAGCTTCACG | |
| rbcL | ES-rbcL-628F | Forward |
CCATTYATGCGTTGGAGAGATCG | |
| rbcL | ES-rbcL-654R | Reverse |
GAARCGATCTCTCCAACGCAT | |
| rps4 | rps5 | Forward | ATGTCCCGTTATCGAGGACCT | |
| rps4 | trnS | Reverse | TACCGAGGGTTCGAATC |
† Primer used only for sequencing.
Amplifications were visualized using standard gel electrophoresis and imaging approaches. Unincorporated nucleotides and primers were removed from successful reactions by adding 1.0 µl Shrimp Alkaline Phosphatase (1 unit/µl) and 0.5 µl Exonuclease I (10 units/µl) to each reaction and incubating at 37 °C for 15 min. Reactions were then heated to 80 °C for 15 min to inactivate the enzymes.
Sequencing reactions were carried out, in both directions, with the amplification primers, following a standard protocol (
Sequencing reads were independently (for each PCR product) assembled and edited using Sequencher (Gene Codes Corporation). The 110 new consensus sequences were added to the Fern Lab Database (http://fernlab.biology.duke.edu) and deposited into GenBank (Table 1). For four (of thirty-eight) collections, we could only obtain three of the four gene regions targeted (Table 1). For six collections, an atpA and/or rbcL sequence had already been published; these existing sequences (from
Details for the alignments analyzed in this study supporting the recognition of Tryonia.
| Characters | Data | Bipartitions | ||||
|---|---|---|---|---|---|---|
| Dataset | Taxa | Total | Included | Variable | Missing |
Supported |
| atpA | 30 | 1506 | 629 | 113 | 1.04% | 11 |
| chlL | 31 | 523 | 523 | 120 | 0.92% | 15 |
| rbcL | 31 | 1309 | 1309 | 250 | 0.39% | 15 |
| rps4 | 35 | 1176 | 560 | 177 | 1.77% | 17 |
| Combined | 38 | 4514 | 3021 | 660 | 17.76% | 25 |
† Calculation based on included characters
‡ Bayesian posterior probability ≥ 0.95
Bayesian phylogenetic analyses were conducted independently for each of the four single-gene datasets using MRBAYES version 3.2.1 (
We compared the results of our single-gene analyses, looking for conflicts that were supported by a Bayesian posterior probability ≥ 0.95. Finding none, we concatenated the four datasets. The resulting 38-taxon combined dataset was analyzed as above, but with model parameters estimated and optimized separately for each gene and each run proceeding for 20 million generations. We sampled trees every 16, 000 generations and excluded the first four million generations from each run prior to calculating a majority-rule consensus phylogeny with clade posterior probabilities.
The four single-gene (atpA, chlL, rbcL, and rps4) datasets contained varying amounts of phylogenetic signal, providing significant support (Bayesian posterior probability, BPP ≥ 0.95) for as few as 11 and as many as 17 bipartitions (Table 3). The single-gene trees were largely consistent in their resolved relationships (trees not shown) and there were no well-supported (BPP ≥ 0.95) conflicts among them.
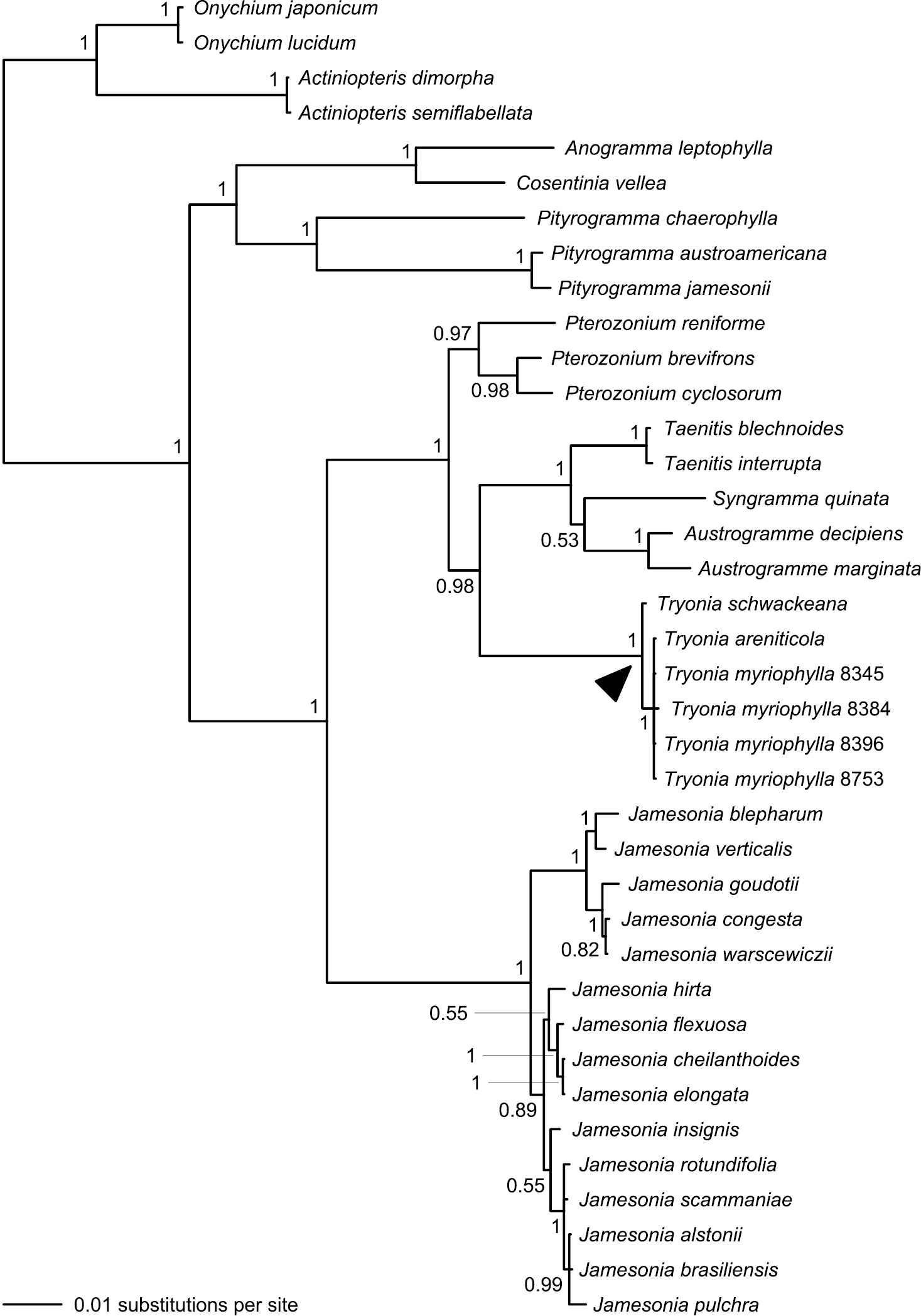
Our combined four-gene dataset comprised a total of 4514 characters, of which 660 were variable (Table 3). Analysis of this dataset resulted in a phylogeny with considerably improved support relative to the single-gene phylogenies; 25 bipartitions had a BPP ≥ 0.95 (Fig. 5). The separation of Actiniopteris and Onychium from the remaining taenitidoid genera was well supported (BPP = 1.00). Anogramma, Cosentinia, and Pityrogramma formed a well-supported clade that was, in turn, well-supported as sister to a robust clade including Austrogramme, Pterozonium, Syngramma, Taenitis, and all sampled species previously assigned to either Jamesonia or Eriosorus (Fig. 5).
Phylogeny resulting from Bayesian analysis of our combined four-gene (atpA, chlL, rbcL, and rps4) plastid dataset. Posterior probabilities (≥ 0.50) are provided at the nodes. Note that species now treated in Tryonia (black arrowhead) are distinct from Jamesonia, the genus in which these species were most recently placed. Numbers provided for Tryonia myriophylla samples are Fern Lab Database voucher numbers (Table 1).
The vast majority of our Jamesonia sensu lato collections come together in a clade on a rather long branch; within this clade branches are short and support is frequently lacking. Six samples previously included within Jamesonia sensu lato are not allied to that larger clade, but rather are embedded within a well-supported clade that also contains Austrogramme, Pterozonium, Syngramma, and Taenitis (Fig. 5).
Most species previously assigned to Eriosorus and Jamesonia sensu stricto have been consistently resolved together in a well-supported clade (
Eriosorus myriophyllus was shown by
In her monograph of Eriosorus,
Based on our current dataset, we do not consider the precise phylogenetic position of Tryonia (within the Austrogramme, Pterozonium, Syngramma, Taenitis, and Tryonia clade) to be fully resolved. Although our combined analysis clearly places Tryonia sister to Austrogramme, Syngramma, and Taenitis (collectively), this relationship is not well supported in any single-gene analysis. The atpA and rbcL datasets do place Tryonia sister to Taenitis (atpA and rbcL sequences were not available for Austrogramme and Syngramma), but support is lacking (BPP = 0.61 and 0.83, respectively). Likewise, the rps4 dataset resolves Tryonia as sister to Austrogramme, Syngramma, and Taenitis without significant support (BPP = 0.88). Strong single-gene support for the precise position of Tryonia only comes from the chlL dataset, where Tryonia is most closely related to Pterozonium (BPP = 1.00).
Two of the species of Tryonia included in our phylogenetic analysis (Tryonia areniticola and Tryonia schwackeana) are endemic to Brazil; the third sampled species (Tryonia myriophylla) also occurs in Uruguay, near its border with the Brazilian state of Rio Grande do Sul. Although the Andes are the center of diversity for Jamesonia (as newly circumscribed herein), this genus is not entirely geographically distinct from Tryonia. In the recently published Catálogo de Plantas e Fungos do Brasil, a total of nine species are ascribed to Eriosorus or Jamesonia (
urn:lsid:ipni.org:names:77136217-1
http://species-id.net/wiki/Tryonia
Figs 4, 6–9Similar to some species of Jamesonia, but with stramineous rather than castaneous rachises.
Tryonia myriophylla (Sw.) Schuettp., J.Prado & A.T.Cochran, comb. nov., Gymnogramma myriophylla Sw., Kongl. Vetensk. Acad. Handl. 1817(1): 58. 1817.
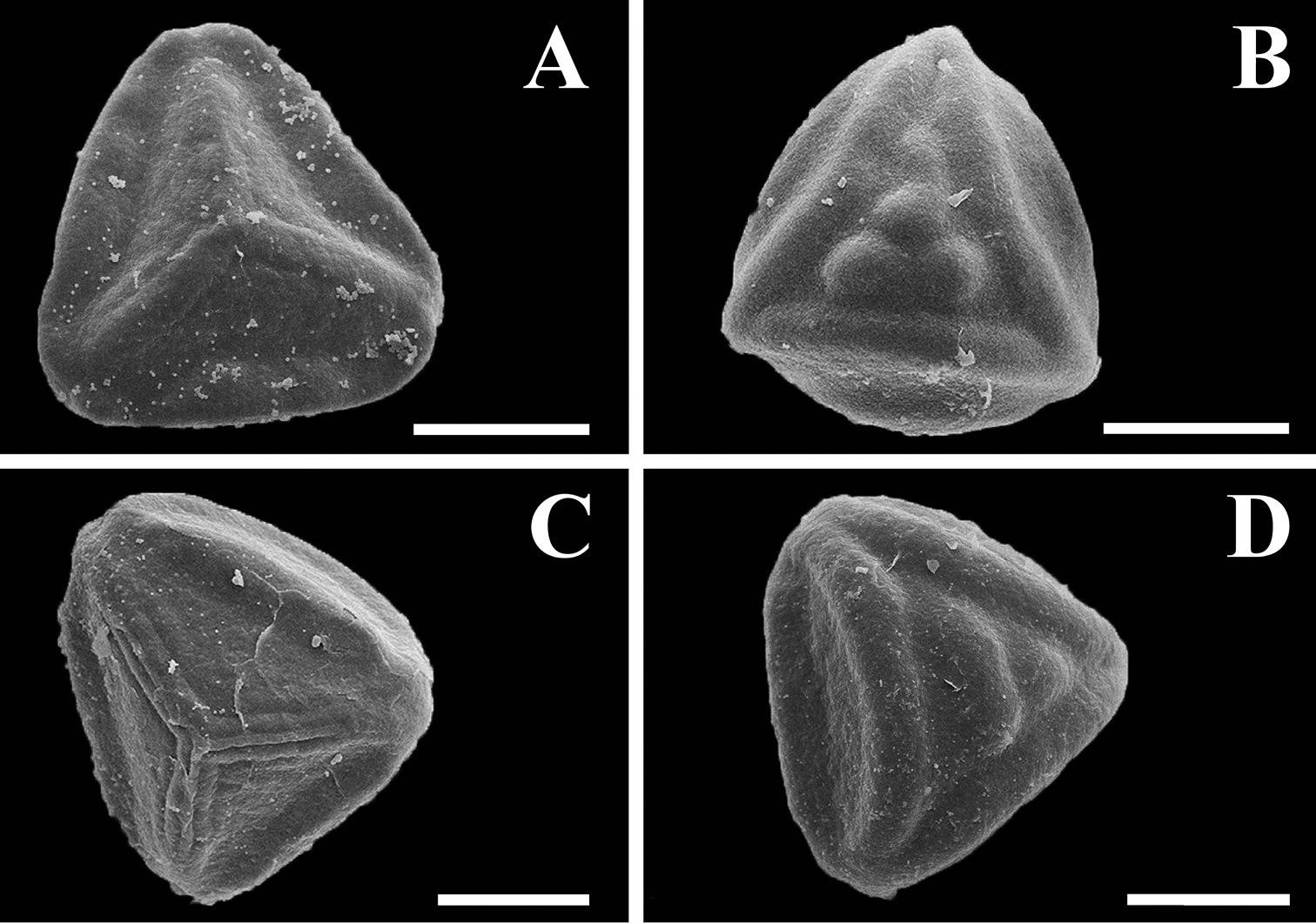
Plants terrestrial, rupicolous, or saxicolous. Rhizomes creeping to erect at apex, compact, with appressed hairs or crispate bristles, sometimes rigid, ruddy brown, darker at the base. Fronds erect, 6–100 cm long; petioles terete or sulcate adaxially, brown at base and stramineous distally, from 1/8 as long to equal the length of the lamina, densely to sparsely pubescent, the hairs short and erect or long and crispate, hyaline or reddish brown at the cell junctions, glandular or non-glandular; laminae linear to elongate-triangular, 1 or 2-pinnate-pinnatissect to 1–3-pinnate-pinnatifid, 4.0–48 cm long, 1.0–14 cm wide, determinate; rachises straight, sometimes slightly flexuous, terete or sulcate adaxially, stramineous, pubescent, the hairs like those of the petioles; pinnae ascending to patent to the rachis, oblong to deltate, 0.5–10 cm long, 0.5–5 cm wide, membranaceous to herbaceous, densely to sparsely pubescent on both surfaces, the hairs glandular, hyaline or with the terminal cell light to dark reddish brown, 2–5-celled, or hairs non-glandular, hyaline or reddish brown at the cell junctions, 2–5(–7)-celled; ultimate segments entire and round or emarginate; veins free. Sporangia borne along the veins, short-stalked, stalks 1–2-celled, stomia with 2–4 indurated cells; spores trilete, tetrahedral-globose, with an equatorial flange, distal face coarsely tuberculate, proximal face with prominent ridges, brown, 40–60 µm (Fig. 9).
Tryonia areniticola (Schwartsb. & Labiak) Schuettp., J.Prado & A.T.Cochran. Schwartsburd 487 (SP), inset detail of (stramineous) rachis magnified 4×.
Tryonia schwackeana (Christ) Schuettp., J.Prado & A.T.Cochran. Schuettpelz 1433 (MO), inset detail of (stramineous) rachis magnified 4×. Image modified from http://www.tropicos.org/Image/100140486.
Tryonia sellowiana (Kuhn) Schuettp., J.Prado & A.T.Cochran. Mulford 710 (US), inset detail of (stramineous) rachis magnified 4×.
Spores of Tryonia. A. Tryonia myriophylla proximal view, Wacket s.n. (US) B Tryonia myriophylla distal view, Wacket s.n. (US) C Tryonia areniticola proximal view, Kummrow 2773 (US) D Tryonia areniticola distal view, Kummrow 2773 (US). All scale bars are 20 µm.
The generic name honors Dr. Alice Faber Tryon, who made extraordinary contributions to fern systematics and published taxonomic revisions of both Jamesonia sensu stricto and Eriosorus (from which Tryonia is segregated herein).
Tryonia occurs primarily in southeastern Brazil. However, one species (Tryonia myriophylla) can also be found in Uruguay (Cerro Largo: Sierra Souza), near the Brazilian border. The genus is mostly restricted to the Atlantic Forest, along shaded streams, on damp shaded sandstone, or in more open places (but here shaded by shrubs); 600–2300 m.
Tryonia can be distinguished most readily from Jamesonia by its stramineous rachises, but its gross morphology is also reasonably distinct.
Tryonia comprises the following species.
urn:lsid:ipni.org:names:77136218-1
http://species-id.net/wiki/Tryonia_areniticola
Figs 6, 9Eriosorus areniticola Schwartsb. & Labiak (Amer. Fern J. 98: 160. 2008).
Brazil: Paraná: Jaguariaíva: Parque Estadual do Cerrado, 12 April 1994, P.H. Labiak 182 (holotype: UPCB; isotypes: SP!, UC).
Brazil: Paraná, Rio Grande do Sul, Santa Catarina (probably), and São Paulo.
Based on the gene regions included in our analysis, we found Tryonia areniticola to be genetically indistinguishable from Tryonia myriophylla, despite the presence of several morphological differences (
urn:lsid:ipni.org:names:77136219-1
http://species-id.net/wiki/Tryonia_myriophylla
Figs 4, 9Gymnogramma myriophylla Sw. (Kongl. Vetensk. Acad. Handl. 1817(1): 58. 1817).
Brazil: [Minas Gerais]: Villa Rica [now Ouro Preto], Aug 1815, G.W. Freyriss s.n. (lectotype [designated by Tryon, 1970]: S-R-2467, image!; isolectotypes: BM 000936677, image!, S-R-2469, image!).
Brazil: Bahia, Espírito Santo, Minas Gerais, Paraná, Rio de Janeiro, Santa Catarina, São Paulo, and Rio Grande do Sul. Uruguay: Cerro Largo.
urn:lsid:ipni.org:names:77136220-1
http://species-id.net/wiki/Tryonia_schwackeana
Fig. 7Gymnogramma schwackeana Christ in Schwacke (Pl. Nov. Mineiras 2.18. 1900).
Brazil: [Minas Gerais]: Ouro Preto, C.A.W. Schwacke 9389 (lectotype [designated by Tryon, 1970]: P 00603566, image!; isolectotype: GH 00021287, image!).
Brazil: Bahia and Minas Gerais.
urn:lsid:ipni.org:names:77136221-1
http://species-id.net/wiki/Tryonia_sellowiana
Fig. 8Gymnogramma sellowiana Mett. ex Kuhn (Linnaea 36:69. 1869).
Brazil, Sello 1365 (lectotype [designated by Tryon, 1970]: B-Herb. Mett., image!; isolectotype: B, image!)
Brazil: Minas Gerais.
This research was funded in part by the National Science Foundation (NSF awards DEB-0717398, DEB-0717430, DEB-1145614, and DEB-1145925), the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq award 301157/2010-3), and a Cahill Award from the University of North Carolina Wilmington. Amanda Grusz, Anders Larsson, Robbin Moran, Carl Rothfels, Harald Schneider, Alan Smith, Michael Sundue, and George Yatskievych provided material or assisted in material acquisition. Alex Davila, Patricia Kelley, Jerald Pinson, Ann Stapleton, Marcel van Tuinen, and three reviewers provided helpful comments on the manuscript. Specimen images were provided by the Missouri Botanical Garden and by Ingrid Lin of the Smithsonian Institution. Scott Whittaker assisted with spore imaging and Regina Hirai prepared the spore plate.